× close
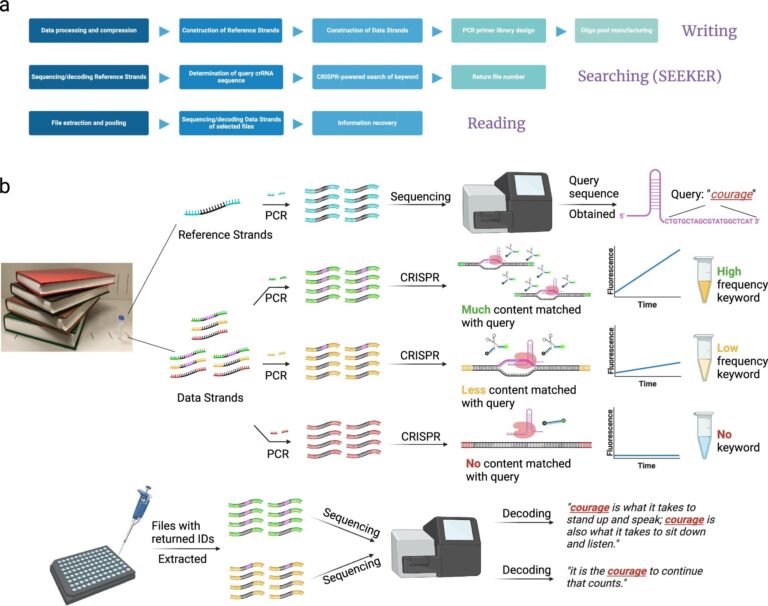
A framework for DNA data storage systems with search capabilities and the general workflow of SEEKER. be The complete framework for a searchable DNA data storage system includes writing, searching, and reading data. b The oligo pool that stores text data is constructed in two parts: a reference strand and a data strand. The reference strand is typically composed of 100-200 oligos and can be used as a dictionary used to map the data strand to a binary code or pre-sequenced to determine the crRNA spacer sequence for the query of interest. can. For example, the keyword “courage” corresponds to the crRNA sequence “CTGTGCTAGCGTATGGCTCAT”. The data strands are selectively amplified according to the file ID and incubated with the Cas12a-crRNA ribonucleoprotein complex. If the amplified file contains many repetitions of the keyword “courage”, the fluorescence intensity will increase rapidly, producing a strong fluorescence signal within a short time. When fewer instances of the keyword “courage” appear in the file, the fluorescence enhancement is delayed and the endpoint fluorescence intensity becomes weaker. If the keyword “courage” is not found in the file, no fluorescence will be detected. After searching, files that generate a positive signal are recognized as containing the desired data and undergo next-generation sequencing to restore their full content. In this example, two occurrences of the keyword “courage” will generate a stronger signal, and one occurrence of the keyword “he” will generate a weaker signal. Illustration created by BioRender.com. credit: nature communications (2024). DOI: 10.1038/s41467-024-46767-x
The digital age has led to an explosion of data of all kinds. Traditional data storage methods such as hard drives are starting to face challenges due to limited storage capacity. As the demand for data storage increases, so does the popularity and need for alternative media for data storage.
DNA is one of the emerging solutions for storing data due to its physical density, data longevity, and data encryption capabilities. Any information that can be stored on a hard drive can also be converted into DNA sequences, including text, images, audio, and movies.
However, although DNA is a promising solution to meet data storage needs, performing searches within DNA strands can be cumbersome and difficult.
“Archiving information on synthetic DNA has emerged as an attractive solution to address the explosion of data in modern society. However, quantitatively querying data stored on DNA is It remains a challenge,” said Changchun Liu, professor in the Department of Biomedical Sciences. Engineering at UConn Health.
in nature communicationsLiu and a team of researchers used a quantitative search engine powered by clustered regularly interspaced short palindromic repeats (CRISPR) to easily and effectively search data stored in DNA. I found a way to search for it.
In his paper, Liu introduces Search Enablement through Enzymatic Keyword Recognition (SEEKER), which utilizes CRISPR-Cas12a to quantitatively identify keywords in files stored in DNA.
“DNA is a promising medium for data storage because of its stability and high information density. Theoretically, one gram of DNA can store 215 petabytes of data, which is approximately 100 million It’s the size of a movie. It’s like a hard drive that stores information in binary data.” DNA consists of four molecules: adenine (A), thymine (T), cytosine (C), and guanine (G). Stores information in sequences of nucleobases.
“With advances in DNA synthesis technology and next-generation sequencing, preservation of DNA data is becoming a reality,” explains Jiongyu Zhang, a graduate student in Liu’s lab and first author of the paper.
Liu leveraged his expertise in CRISPR technology to devise a better solution for searching within DNA strands.
CRISPR is an adaptive immune mechanism that can identify specific infectious DNA sequences in cells overwhelmed with interfering genes, similar to keyword searches in databases.
SEEKER utilizes CRISPR to rapidly generate visible fluorescence or light when a DNA target corresponding to the keyword of interest is present. SEEKER can successfully perform quantitative text searches because the rate of increase in fluorescence intensity is proportional to the keyword frequency.
In this paper, the researchers were able to identify keywords in 40 files against the backdrop of approximately 8,000 unrelated terms.
“Overall, SEEKER provides a quantitative approach to perform parallel searches, including metadata searches, against the complete content stored in DNA with easy implementation and rapid result generation,” said Liu. he explains.
For more information:
Jiongyu Zhang et al., Quantitative Keyword Search Engine Using CRISPR in DNA Data Storage; nature communications (2024). DOI: 10.1038/s41467-024-46767-x